Projects
P1: Biosynthesis and utilisation of SAM diastereomers as tools for the characterisation of SAM-dependent enzymes and product diversification

S-Adenosyl-L-methionine (SAM) is one of the most versatile cofactors and is involved in a remarkably wide range of reaction types in almost all life forms. The most prominent function of SAM is to serve as a methyl group donor for methyltransferases. However, all substituents at the sulfonium ion are involved in various SAM-dependent enzymatic reactions. Recent advances in the fields of cofactor regeneration and enzymatic synthesis of cofactor analogues, together with the versatile reactivity of SAM makes SAM-dependent enzymes promising tools for various biocatalytic applications. The goal of this project is the enzymatic synthesis of SAM analogues to expand the knowledge on structures, functions, and mechanisms of SAM utilising enzymes as well as enzymes involved in the downstream metabolism of SAM-derived products. The knowledge gained will be translated into efficient and flexible in vitro applications.
Agnes Bartels,
Lars-Hendrik Köppl,
Lukas Gericke,
Prof. Dr. Jennifer Andexer, Freiburg
P2: Structural basis and mechanism of SAM-utilizing PLP-dependent enzymes

S-adenosylmethionine (SAM)-utilizing pyridoxal phosphate (PLP)-dependent enzymes (SAM-PLP enzymes) are an emerging family of biocatalysts that stand out in terms of their remarkable chemical versatility rooted in the combined and potentiated reactivities of the carbanion-stabilizing cofactor PLP and the multifunctional nature of the sulfonium-bearing SAM. Reported enzyme-mediated transformations range from intramolecular cyclopropylations, to intermolecular Claisen-like reactions, C-N bond-forming aza-Michael-type additions, or tandem C-C bond formations via (3+2)-cycloaddition. The steadily increasing number of available genomic and metagenomic data represents a vast resource for the discovery of novel biocatalysts by targeted data mining. However, due to the current lack of in-depth structural and mechanistic knowledge for SAM-PLP enzymes, no methods are available for functional annotation based on sequence data. The goal of the project is to elucidate key structural and mechanistic determinants for SAM binding and conversion by SAM-PLP enzymes and the subsequent development of bioinformatic tools for targeted enzyme discovery approaches by genomic data mining. The results will provide a deeper understanding of the enzymological and evolutionary functions of SAM and will lay the basis to explore and exploit the versatile group of SAM-PLP biocatalysts.
Katrin Zoller,
Tenure-Track-Prof. Dr. Lena Barra, Konstanz
P3: Mechanism and scope of azetidine-1-carboxylic acid synthases
Azetidine-1-carboxylic acid (AZE) has been found to be a non-canonical α-amino acid that is incorporated into microbial secondary metabolites such as vioprolides and azetidomonamides. These metabolites are reported to possess cytotoxic effects and to regulate quorum sensing; therefore, the compounds containing AZE are of interest for their pharmaceutical application. AZE is generated from S-adenosylmethionine (SAM) by the specialized AZE synthases, which are structurally and functionally similar to class-1 methyltransferases. Despite their importance, the detailed mechanisms by which these enzymes catalyze the formation of the strained azetidine ring remain largely unknown. This project aims to reveal the fundamental catalytic principles of AZE synthases using advanced biochemical and structural analysis. It also plans to determine their biochemical potential and find new enzymatic functions from their larger protein family.
Marlena Bolz,
Prof. Dr. Wulf Blankenfeldt, Braunschweig
P4: SAM- and cobalamin- dependent conversion of estrogens into androgens

Bacterial degradation of endocrine disrupting and carcinogenic estrogens is essential for their elimination from the environment. In the anaerobic denitrifying betaproteobacterium Denitratisoma (D.) oestradiolicum, estrogen degradation is initiated by S-adenosylmethionine (SAM)- and electron donor-dependent methylation of ring A of 17β-estradiol to dearomatized 1-dehydrotestosterone. The reaction is catalyzed by an unusual SAM/cobalamin-dependent estrogen methyltransferase (Emt), probably composed of the four subunits of EmtABCD. During estrogen degradation in D. oestradiolicum, C1 metabolism is strongly up-regulated to regenerate SAM (Jacoby et al., 2020). The aim of this project is to isolate and biochemically, kinetically and structurally characterize the SAM/cobalamin-dependent methyltransferase complex that converts estrogens to androgens. SAM analogues provided by collaborating partners within the FOR will be used to define the substrate spectrum of the methyltransferase complex and to generate novel androgen steroids in bioorthogonal approaches. Finally, a whole-cell system based on D. oestradiolicum will be developed to direct C1 metabolism towards SAM regeneration.
Tingyi Zhan,
Prof. Dr. Matthias Boll, Freiburg
Jacoby, C.; Krull, J.; Andexer, J.; Jehmlich, N.; von Bergen, M.; & Brüls, T.; Boll, M. Channeling C1 Metabolism toward S -Adenosylmethionine-Dependent Conversion of Estrogens to Androgens in Estrogen-Degrading Bacteria. mBio 2020, 11, e01259-20 doi: 10.1128/mBio.01259-20.
P5: Exploring the potential of engineered enzyme families for selective N-alkylation of heteroarenes: A convergent synthesis approach with SAM analogs as intermediates
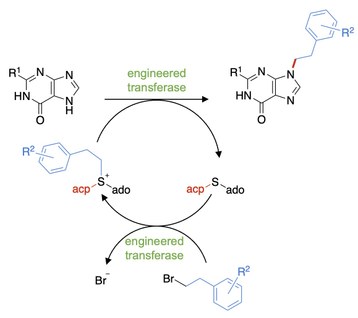
The selective N-alkylation of heteroarenes could drastically shortcut synthesis of complex molecules, especially if two larger fragments could be coupled by selective C-N bond formation in a convergent approach. Here we propose to engineer, understand, and apply enzyme families that synthesize and use S-adenosyl-L-methionine (SAM) analogs as intermediates for selective N-alkylation of heteroarenes. Our goal is to generate two enzyme families, namely sulfonium ion synthases that generate SAM analogs from S-adenosyl-L-homocysteine and "off the shelf" haloalkanes, and N-alkyltransferases that use these SAM analogs as co-substrates in selective C–N bond formations with heteroarene building blocks. This research i) aims to develop a catalytic method for a sought-after chemical transformation, ii) uses in silico mutagenesis for computational design of mutant libraries to systematically explore large regions of the amino acid sequence space, and iii) explores the potential of SAM analogs in convergent synthesis by coupling readily available fragments to complex molecules
Kai Schülke,
Marius Schnutenhaus,
Jun.-Prof. Dr. Stephan Hammer, Bielefeld
P6: Characterisation and engineering of the cobalamin-dependent Radical SAM methyltransferase Orf29 for the synthesis of novel SAM derivatives

The unusual, non-proteinogenic and cyclopropane-containing amino acid (1S,2S)-2-methyl-1-aminocyclopropane-1-carboxylic acid ((1S,2S)-MeACC) is biosynthesised in a two-step enzymatic process that utilises S-adenosyl-L-methionine (SAM) as initial substrate. In the first step, a cobalamin-dependent Radical SAM methyltransferase (B12-RSMT), termed Orf29, catalyses the methylation of the methionine moiety of SAM yielding (4′′R)-4′′-methyl-SAM (1). Second, a cyclisation reaction is leading to formation of (1S,2S)-MeACC (1). This product serves as a building block of pharmaceutically relevant substances exhibiting antibiotic and/or antitumor activities (2,3).
In this project, the B12-RSMT Orf29 will be characterised in order to elucidate the structure-function relationship of this intriguing enzyme, which utilises SAM in three distinct roles: as cofactor for initiation of radical catalysis, as methyl group donor and as substrate. In order to understand the catalytic mechanism of Orf29, we aim to elucidate the sequence of individual reaction steps and define the enzyme active site. Besides the functional and structural characterisation of Orf29, we aim to engineer this enzyme to a versatile platform for the synthesis of diverse SAM derivatives for later incorporation into novel, modified peptide antibiotics.
Fabian Piskol,
Prof. Dr. Gunhild Layer, Freiburg
(1) Maruyama, C.; Chinone, Y.; Sato, S.; Kudo, F.; Ohsawa, K.; Kubota, J.; Hashimoto, J.; Kozone, I.; Doi, T.; Shin-Ya, K.; Eguchi, T.; Hamano, Y. C-Methylation of S-adenosyl-L-methionine occurs prior to cyclopropanation in the biosynthesis of 1-amino-2-methylcyclopropanecarboxylic acid (norcoronamic acid) in a bacterium. Biomolecules 2020, 10 (5), 775. DOI: 10.3390/biom10050775.
(2) Kurosawa, K.; Takahashi, K.; Tsuda, E. SW-163C and E, novel antitumor depsipeptides produced by Streptomyces sp. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot 2001, 54 (8), 615–621. DOI: 10.7164/antibiotics.54.615.
(3) Nakaya, M.; Oguri, H.; Takahashi, K.; Fukushi, E.; Watanabe, K.; Oikawa, H. Relative and absolute configuration of antitumor agent SW-163D. Biosci Biotechnol Biochem 2007, 71 (12), 2969–2976. DOI: 10.1271/bbb.70371.
P7: Dearomatisation by asymmetric methylation

S-Adenosyl-L-methionine (SAM)-dependent methyltransferases (MTs) can methylate various nucleophiles in an SN2 reaction and are involved in the methylation of a variety of natural products. Compared to methylation on polarizable heteroatoms, C-methylation requires activation of the carbon atom of the substrates by an adjacent functional group to form a nucleophilic intermediate and allow nucleophilic attack on the methyl moiety of SAM. Exemplary substrates are enolizable ketones or phenolic compounds. In this project, we focus on the SAM-dependent C-methylation of α-keto acids and aromatic substrates. The structural and mechanistic requirements for regio-, chemo-, and stereoselective methylation reactions are investigated. These approaches enable novel asymmetric biocatalytic transformations to chiral compounds.
Juliane Breiltgens,
Ziruo Zou,
Prof. Dr. Michael Müller, Freiburg
P8: Integration of sequence and reaction data for the design and engineering of methionine adenosyltransferases and other SAM-dependent enzymes



An integrated computational platform for analysing data on sequence, structure, and function of enzymes will be developed and applied to design tailored methionine adenosyltransferases (MAT) with an altered substrate profile. In cells, MATs produce S‑adenosylmethionine (SAM) from 5´-adenosine triphosphate (ATP) and L-methionine. The target for the envisioned enzyme variants is the control of the enzyme’s discrimination of L- and D‑methionine. In addition to selective variants, also unselective variants will be designed and characterised. The integrated platform will be developed alongside the MAT project and comprises bioinformatics workflows and a research data management toolbox for biocatalytic data. The bioinformatics workflows are applied for studying sequence-function relationships, finding new enzyme candidates in sequence databases, and designing highly enriched mutant libraries. The research data management toolbox is based on the standardised data exchange format EnzymeML and is applied for managing, analysing, and publishing experimental and modelling results according to the FAIR data principles. The platform development is a collaborative project using existing tools such as GitHub, Jupyter, the Biopython library, and Galaxy, and incorporates existing databases, formats, and standards. The platform is used by all FOR 5596 partners, who will be enabled to install it locally, adapt it to their needs, and apply if for analysing and publishing their data and for sharing methods and results. The computational tools developed in P8 are reusable and extensible, the workflows enable reproducibility of data analysis, and the use of standardised formats make results interoperable. The tools developed in P8 focus to the needs of FOR 5596, but can be extended and generalised beyond FOR 5596 and thus contribute to the digitalisation of (bio)catalytic sciences.
Max Häußler,
Prof. Dr. Jürgen Pleiss, Stuttgart,
Prof. Dr. Jennifer Andexer, Freiburg
P9: Enzymatic generation of double-modified SAM analogues
Methyltransferases for biomolecular labelling
Exploiting nature's bioalkylation systems has become common for labelling biomolecules via transfer of non-natural groups. Methyltransferases (MTases) utilize S-adenosyl-L-methionine (AdoMet) as co-substrate to methylate a diverse range of DNA, RNA, protein, or small molecule substrates. Non-natural AdoMet analogues with modifications at the sulfonium centre have proven useful for the transfer of various functional groups, including photocleavable or clickable ones. If a promiscuous MTase is available, this approach proves powerful for site-specific labelling of a substrate with a group of interest, enabling downstream applications, such as enrichment or modulations of the biological function.
Cascade reactions and metabolic labelling approaches
Methionine adenosyltransferases (MATs) use ATP and methionine analogues to generate the respective AdoMet analogues. The MAT reaction can be performed in one pot with the MTase as an enzymatic cascade for chemoenzymatic synthesis and site-specific transfer. Methionine analogues as starting point provide two key advantages, namely (i) their high stability (compared to AdoMet analogues that have limited half-lives in aqueous solution) and (ii) their cellular uptake (whereas AdoMet is not cell-permeable). These features are exploited in metabolic labelling approaches, in which cells are treated with methionine analogues, AdoMet analogues are formed inside the cells and used by promiscuous MTases.
Lack of selectivity in complex systems
These approaches have proven powerful to analyse methylation target sites in select biomolecular species (notably RNA) that can be readily separated from other components of the cell. However, the complexity of the living systems brings unique challenges for molecular labelling in vivo. The main concerns would be selectivity and efficiency issues if two or more MTases are present at the same time. The challenge to realize selective MTase-based modifications in complex mixtures or in cells is thus enormous, as there are currently >400 unique MTase entries for Homo sapiens and >60 entries for Escherichia coli in UniProt.
In many MTases, the sulfonium centre of AdoMet is solvent-exposed in the AdoMet binding pocket, enabling many MTases to accept sulfonium centre-modified AdoMet analogues. Consequently, it can be anticipated that metabolic labelling via methionine analogues would lack selectivity. Because of this, in the present project, we are focusing on double-modified AdoMet analogues (DM-AdoMets) to overcome the MTase promiscuity. In the DM-AdoMets, additionally the nucleoside moiety is also modified, where the ligand is less solvent-exposed. Thereby, it is expected that substitution of this moiety may successfully disrupt the interactions with MTases that are not tailored for DM-AdoMets, allowing selective targeting of an engineered MTase of interest.
Mehmet Ergüven,
Prof. Dr. Andrea Rentmeister, Münster
P10: Diversity-oriented biocatalytic production of complex polyamines

In our research we will develop a simple and scalable one-pot synthesis of SAH starting from racemic homocysteine thiolactone and adenosine. This process is catalyzed by recombinant α-amino-ε-caprolactam racemase, bleomycin hydrolase, and SAH hydrolase. The reaction proceeds to completion with near-stoichiometric mixtures of reactants, driven by the irreversible and stereoselective hydrolysis of thiolactone, followed by the thermodynamically favorable condensation of homocysteine with adenosine. We plan to demonstrate that this method can be utilized to supplement preparative methylation reactions with SAH as a cofactor, as well as to synthesize and screen S-nucleosyl homocysteine derivatives in the search for stabilized SAM analogs and efficient substrate of polyamine biosynthetic enzymes.
Dr. Xiaojin Wen,
Prof. Dr. Florian P. Seebeck, Basel